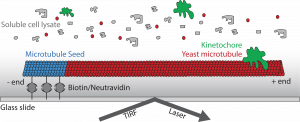
Microtubules are cytoskeletal structures that are necessary for a number of functions within cells, including maintaining cell shape, transporting intracellular cargoes, and segregating DNA during mitosis and meiosis. The assembling of microtubules from αβ tubulin dimers imbues the structure with an inherent polarity which in turn gives rise to the stochastic growing and shrinking of the polymer ends known as dynamic instability. Though tubulin on its own can create microtubules, their growth and shrinkage must be spatially and temporally regulated to coordinate forms that provide function. Microtubule-associated proteins provide this regulation.
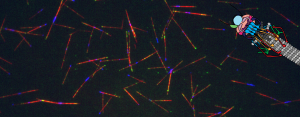
Our lab studies microtubule behavior and function by utilizing the budding yeast, Saccharomyces cerevisiae, because of its relatively simplistic microtubule network, genetic tractability, and amenity to a variety of experimental methods, including biochemistry and microscopy. Recent work has examined the coordination of microtubule activity in the spindle apparatus and their breakdown at the end of mitosis. Currently, we have developed a new assay which allows the observation of single microtubules in lysate similar to in vitro systems, but in total cellular context. This tool will give us further insight into how microtubule-associated proteins affect dynamic instability in order to create functional microtubule structures.
Recent Publications
Torvi, J. R., Wong, J., Serwas, D, Moayed, A, Drubin, D. G., Barnes, G. (2022). Reconstitution of kinetochore motility and microtubule dynamics reveals a role for a kinesin-8 in establishing end-on attachments eLife (2022), https://doi.org/10.7554/eLife.78450
Bergman, Z. J., Wong, J., Drubin, D. G., & Barnes, G. (2018). Microtubule dynamics regulation reconstituted in budding yeast lysates. J Cell Sci, jcs-219386.
Ibarlucea-Benitez, I., Ferro, L. S., Drubin, D. G., & Barnes, G. (2018). Kinesins relocalize the chromosomal passenger complex to the midzone for spindle disassembly. J Cell Biol, jcb-201708114.
Krefman, N. I., Drubin, D. G., & Barnes, G. (2015). Control of the spindle checkpoint by lateral kinetochore attachment and limited Mad1 recruitment. Molecular biology of the cell, 26(14), 2620-2639.
Peng, Y., Moritz, M., Han, X., Giddings, T.H., Lyon, A., Kollman, J., Winey, M., Yates, J., Agard, D.A., Drubin, D.G. and Barnes, G. (2015). Interaction of CK1δ with γTuSC ensures proper microtubule assembly and spindle positioning. Molecular biology of the cell, 26(13), pp.2505-2518.
Pigula, A., Drubin, D. G., & Barnes, G. (2014). Regulation of mitotic spindle disassembly by an environmental stress-sensing pathway in budding yeast. Genetics, 198(3), 1043-1057.