Current and emerging imaging techniques are capable of producing tremendous amounts of quantitative data. Our lab uses several state-of-the-art microscopes and actively collaborates with leaders in the field who are pushing the boundaries of spatial and temporal resolution to generate rich imaging datasets. We use open source and custom written code to extract, clean, and analyze features from these data to elucidate fundamental biological processes.
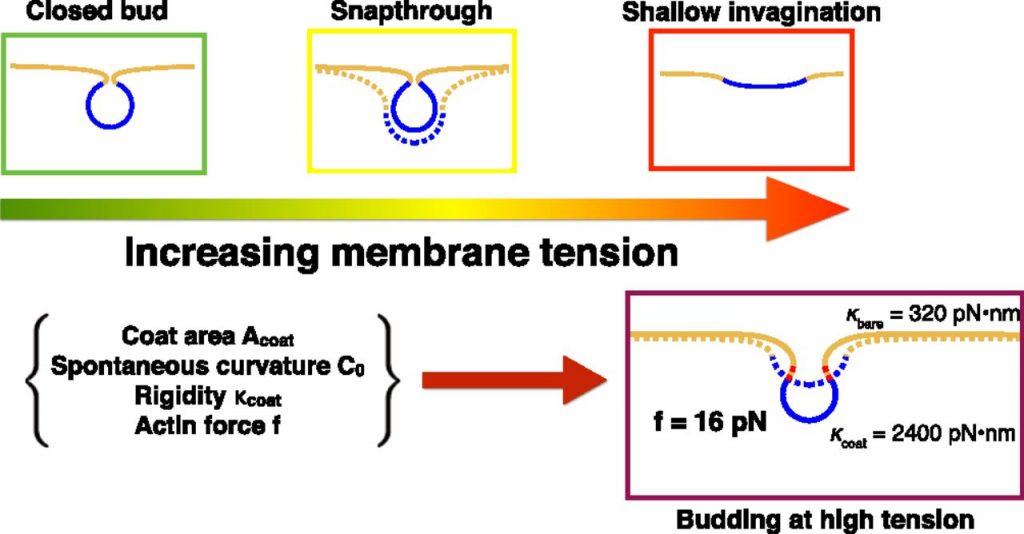
Additionally, our lab has an active collaboration with the Rangamani Lab in which we use mathematical modeling and simulations to understand the mechanochemistry of clathrin-mediated endocytosis (CME) as well as general principles of membrane bending and force generation by the actin cytoskeleton. Recent work has involved modeling the effects of membrane tension on CME in which we proposed general design principles for efficient vesiculation.
Recent publications
Cail, R. C., Shirazinejad, C. R., Drubin, D. G. (2022). Induced nanoscale membrane curvature bypasses the essential endocytic function of clathrin J Cell Biol (2022) 221 (7): e202109013.
Serwas, D., Akamatsu, M., Moayed, A., Vegesna, K., Vasan, R., Hill, J. H., Schöneberg, J., Davies, K., Rangamani, P., Drubin, D. G. (2022). Mechanistic insights into actin force generation during vesicle formation from cryo-electron tomography Dev Cell j.devcel.2022.04.012.
* Gómez-Varela, A. I., Stamov, D. R., Miranda, A., Alves, R., Barata-Antunes, C., Dambournet, D., Drubin, D. G., Paiva, S., De Beule, P. A. A. (2020). Simultaneous Co-Localized Super-Resolution Fluorescence Microscopy and Atomic Force Microscopy: Combined SIM and AFM Platform for the Life Sciences. Scientific Reports, 10 (1), 1122, s41598-020-57885-z.
* Akamatsu, M., Vasan, R., Serwas, D., Ferrin, M. A., Rangamani, P., & Drubin, D. G. (2020). Principles of self-organization and load adaptation by the actin cytoskeleton during clathrin-mediated endocytosis. eLife, 9, e49840.
Schöneberg, J., Dambournet, D., Liu, T. L., Forster, R., Hockemeyer, D., Betzig, E., & Drubin, D. G. (2018). 4D cell biology: big data image analytics and lattice light-sheet imaging reveal dynamics of clathrin-mediated endocytosis in stem cell derived intestinal organoids. Molecular biology of the cell, mbc-E18.
Dambournet, D., Sochacki, K. A., Cheng, A. T., Akamatsu, M., Taraska, J. W., Hockemeyer, D., & Drubin, D. G. (2018). Genome-edited human stem cells expressing fluorescently labeled endocytic markers allow quantitative analysis of clathrin-mediated endocytosis during differentiation. J Cell Biol, jcb-201710084.
Liu, T. L., Upadhyayula, S., Milkie, D. E., Singh, V., Wang, K., Swinburne, I. A., …, Dambournet D., Forster, R., …, Hockemeyer D., Drubin D. G., …, Kirchhausen, T., & Betzig, E. (2018). Observing the cell in its native state: Imaging subcellular dynamics in multicellular organisms. Science, 360(6386), eaaq1392.
Hassinger, J. E., Oster, G., Drubin, D. G., & Rangamani, P. (2017). Design principles for robust vesiculation in clathrin-mediated endocytosis. Proceedings of the National Academy of Sciences, 114(7), E1118-E1127.
[MATLAB code here]
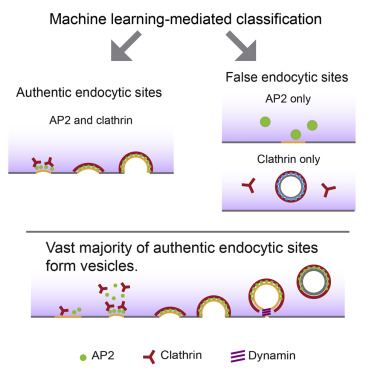
Hong, S. H., Cortesio, C. L., & Drubin, D. G. (2015). Machine-learning-based analysis in genome-edited cells reveals the efficiency of clathrin-mediated endocytosis. Cell reports, 12(12), 2121-2130.