Congratulations to Leanna, David, and Yidi on their collaboration with Margot Riggi and Janet Iwasa to create a molecular animation depicting the full progression of clathrin-mediated endocytosis in budding yeast, along with an accompanying Cell Science at a Glance article and poster. Read the review here.
Au revoir, Paul!
Goodbye Dehbia! We will miss you!
Congratulations to our graduated undergraduates!
Hello is there a doctor in the building?
Well, probably not the kind you need but we do have four new ones in the lab! Congratulations to Cyna, Yui, and Max on becoming doctors*!!! We had an amazing couple days of exit talks, commencement, partying, and family/friend time.
* none of us have actually filed yet, but we promise we will real soon
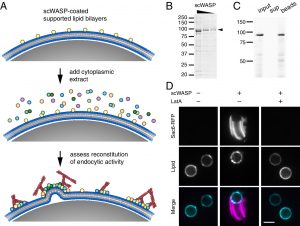
Congratulations to Max and Emily on their new reconstitution paper in PNAS!
The culmination of work from Emily Stoops’ postdoc and Max Ferrin’s PhD was just published as David Drubin’s inaugural article as a new member of the National Academy of Sciences! In the paper they combine time-lapse volumetric confocal fluorescence microscopy with correlative light and electron microscopy to demonstrate the capacity of endocytic proteins to assemble on synthetic membranes and drive vesiculation in a cell-free system. This work was made possible by the excellent shared resources and expertise available to our lab on campus. Most of the fluorescence microscopy was performed on equipment managed by the Molecular Imaging Center, while much of the design and execution of electron microscopy experiments were carried out by Danielle Jorgens in the Electron Microscope Lab. Check out the paper here!
Congratulations to Zane and Jonathan on their new Kar3 paper!
The dynamic duo is at it again, “taking our dynamics reconstitution assay a step further to dissect the differences between the two heterodimers of Kar3, the yeast kinesin-14”! In a heroic effort, Zane and Jonathan have looked at Kar3, Cik1, and Vik1 across the cell cycle in both cells and in our microtubule reconstitution assay. Seriously, it’s a majorly epic undertaking because they don’t just look at these three proteins localization and dynamics, but also their affects on microtubules! I’m so happy for them and their wonderful paper. Check it out here!
Goodbye to Zane & Meiyan
One of the saddest parts of being in a lab, is we become family, but yet people do have to move on with their lives. On one hand, it’s great we end up with friends sprinkled all around the world, but sometimes you just really miss them. And since the last website update, both Zane and Meiyan have moved on to new adventures. Truly, I am so happy for them. Zane and his family have moved down to San Diego were he now is a staff scientist in the Oegema/Desai lab. Finally, his kids can wear shorts all year and have it not being weird and cold! Meiyan and her family are also in warmer climates as they have moved down to Florida, specifically the University of Florida, where her and her husband will continue their research! We all wish them luck, happiness, and success in their new adventures.
Congratulations to Julia and Jonathan for their paper reconstitution kinetochore dynamics in yeast lysate
Major congratulations to them both, but because I (Julia) am writing this, I mainly want to heavily emphasize and thank Jonathan for all his hard work, kindness, patience, and perseverance in working with me. I couldn’t have imaged having a better mentor throughout grad school and I feel like our paper together is something I am truly proud of. It’s our friendship in publication form! Just kidding reader, it’s very much a normal science paper. BUT it is very cool. Check it out if you love microtubule dynamics, kinetochores, reconstitution, motors and especially THE COMBO OF ALL 4! Also I love talking about this data so feel free to reach out anytime if you have a question!
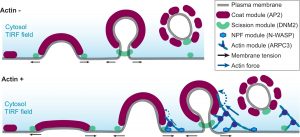
Congratulations to Meiyan and Cyna on their paper regarding asymmetric force production during CME
Congratulations again to Meiyan and Cyna on their beautiful publication and all their hard work. A wonderful collaboration to witness in the lab, Meiyan a stem cell CRISPR editing cell biology wizard and Cyna the statistics quantitative imaging master. Combine the two of them together? Well, read the paper and find out. But, to spoil it a bit, they do beautiful imaging and very thorough quantification to show asymmetrical force production in CME.