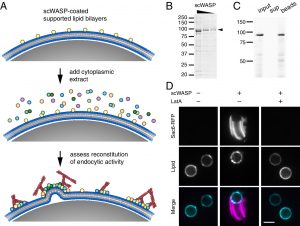
The culmination of work from Emily Stoops’ postdoc and Max Ferrin’s PhD was just published as David Drubin’s inaugural article as a new member of the National Academy of Sciences! In the paper they combine time-lapse volumetric confocal fluorescence microscopy with correlative light and electron microscopy to demonstrate the capacity of endocytic proteins to assemble on synthetic membranes and drive vesiculation in a cell-free system. This work was made possible by the excellent shared resources and expertise available to our lab on campus. Most of the fluorescence microscopy was performed on equipment managed by the Molecular Imaging Center, while much of the design and execution of electron microscopy experiments were carried out by Danielle Jorgens in the Electron Microscope Lab. Check out the paper here!
Category Archives: Papers Published
Congratulations to Zane and Jonathan on their new Kar3 paper!
The dynamic duo is at it again, “taking our dynamics reconstitution assay a step further to dissect the differences between the two heterodimers of Kar3, the yeast kinesin-14”! In a heroic effort, Zane and Jonathan have looked at Kar3, Cik1, and Vik1 across the cell cycle in both cells and in our microtubule reconstitution assay. Seriously, it’s a majorly epic undertaking because they don’t just look at these three proteins localization and dynamics, but also their affects on microtubules! I’m so happy for them and their wonderful paper. Check it out here!
Congratulations to Julia and Jonathan for their paper reconstitution kinetochore dynamics in yeast lysate
Major congratulations to them both, but because I (Julia) am writing this, I mainly want to heavily emphasize and thank Jonathan for all his hard work, kindness, patience, and perseverance in working with me. I couldn’t have imaged having a better mentor throughout grad school and I feel like our paper together is something I am truly proud of. It’s our friendship in publication form! Just kidding reader, it’s very much a normal science paper. BUT it is very cool. Check it out if you love microtubule dynamics, kinetochores, reconstitution, motors and especially THE COMBO OF ALL 4! Also I love talking about this data so feel free to reach out anytime if you have a question!
Congratulations to Meiyan and Cyna on their paper regarding asymmetric force production during CME
Congratulations again to Meiyan and Cyna on their beautiful publication and all their hard work. A wonderful collaboration to witness in the lab, Meiyan a stem cell CRISPR editing cell biology wizard and Cyna the statistics quantitative imaging master. Combine the two of them together? Well, read the paper and find out. But, to spoil it a bit, they do beautiful imaging and very thorough quantification to show asymmetrical force production in CME.
Congratulations to Bob on his paper that discovers a function for clathrin in membrane curvature stabilization in CME
During clathrin-mediated endocytosis (CME), flat plasma membrane is remodeled to produce nanometer-scale vesicles. The mechanisms underlying this remodeling are not completely understood. The ability of clathrin to bind membranes of distinct geometries casts uncertainty on its specific role in curvature generation/stabilization. Here, we used nanopatterning to produce substrates for live-cell imaging, with U-shaped features that bend the ventral plasma membrane of a cell into shapes resembling energetically unfavorable CME intermediates. This induced membrane curvature recruits CME proteins, promoting endocytosis. Upon AP2, FCHo1/2, or clathrin knockdown, CME on flat substrates is severely diminished. However, induced membrane curvature recruits CME proteins in the absence of FCHo1/2 or clathrin and rescues CME dynamics/cargo uptake after clathrin (but not AP2 or FCHo1/2) knockdown. Induced membrane curvature enhances CME protein recruitment upon branched actin assembly inhibition under elevated membrane tension. These data establish that membrane curvature assists in CME nucleation and that the essential function of clathrin during CME is to facilitate curvature evolution, rather than scaffold protein recruitment.
Congratulations to Daniel and co-authors on their cryo-ET study of actin-based force generation in CME!
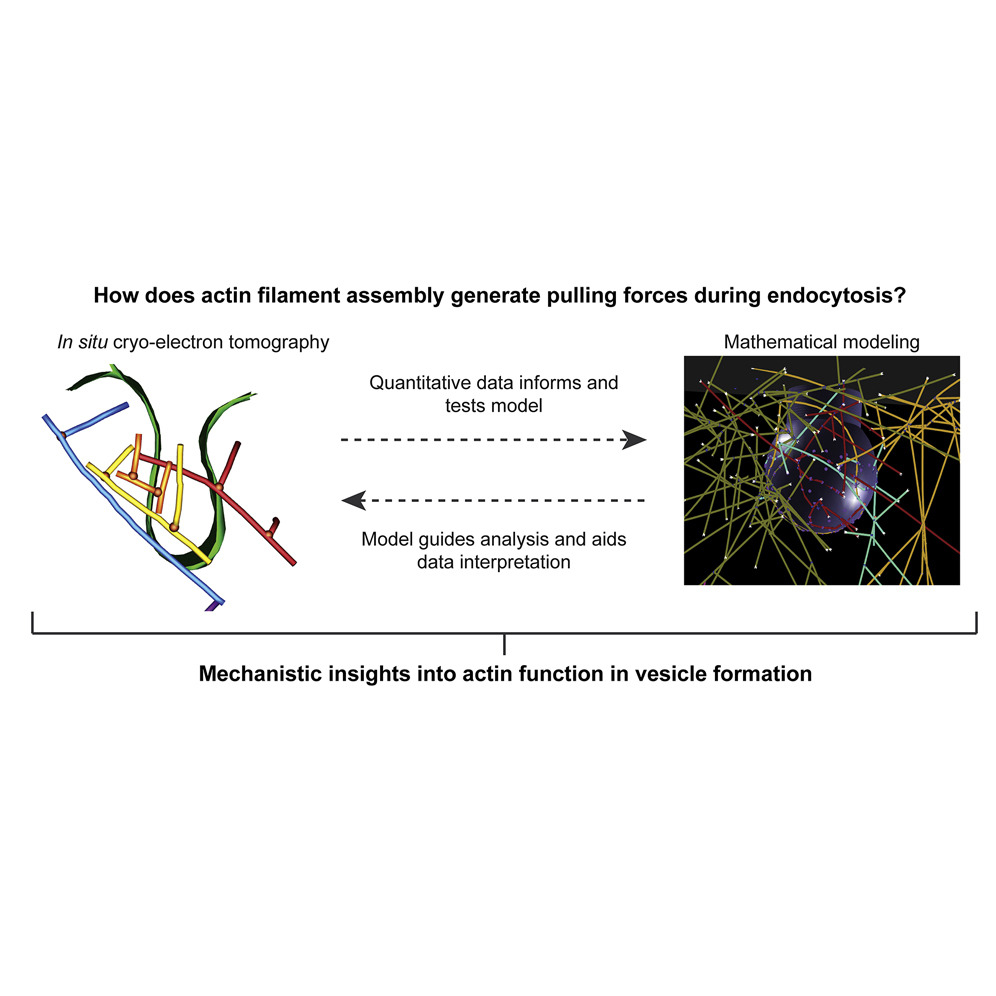
“How does actin filament assembly generate pulling forces during endocytosis?”
Actin assembly provides force for a multitude of cellular processes. Compared to actin-assembly-based force production during cell migration, relatively little is understood about how actin assembly generates pulling forces for vesicle formation. Here, cryo-electron tomography identified actin filament number, organization, and orientation during clathrin-mediated endocytosis in human SK-MEL-2 cells, showing that force generation is robust despite variance in network organization. Actin dynamics simulations incorporating a measured branch angle indicate that sufficient force to drive membrane internalization is generated through polymerization and that assembly is triggered from ∼4 founding “mother” filaments, consistent with tomography data. Hip1R actin filament anchoring points are present along the entire endocytic invagination, where simulations show that it is key to pulling force generation, and along the neck, where it targets filament growth and makes internalization more robust. Actin organization described here allowed direct translation of structure to mechanism with broad implications for other actin-driven processes.
Paper by Ross, Julian, and Paul on endocytic maturation published in JCB
Congratulations to Ross, Julian, and Paul on their paper “Spatial regulation of clathrin-mediated endocytosis through position-dependent site maturation” now published in JCB.
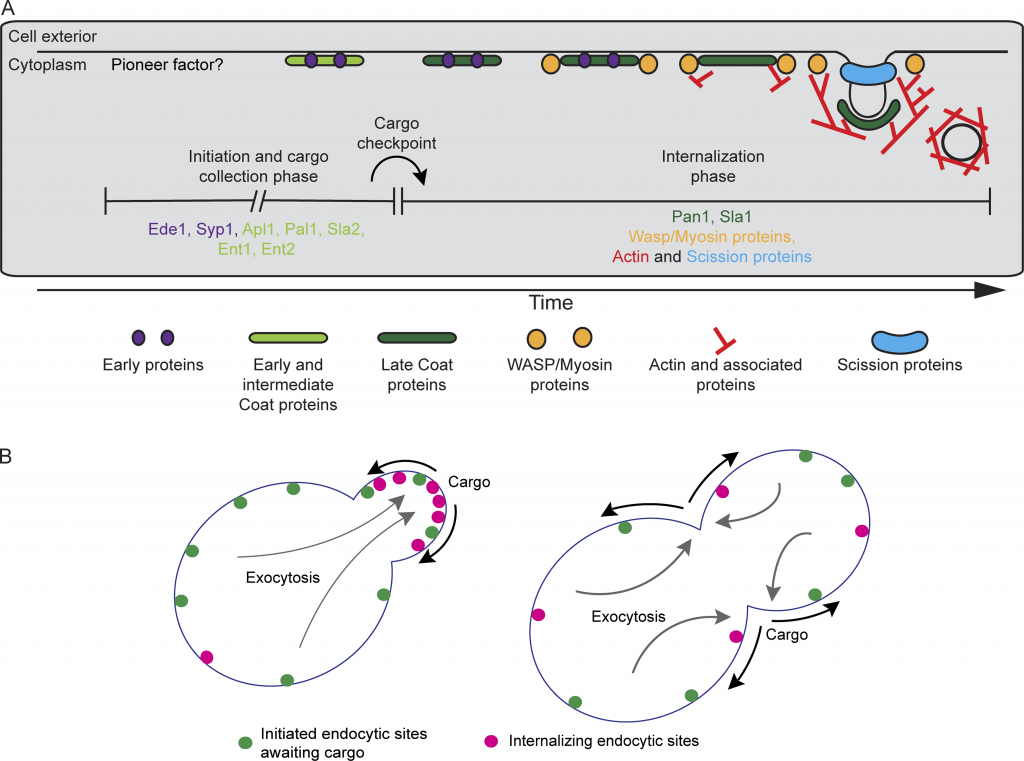
Cargo regulates the transition between two quantitatively distinguishable phases of endocytosis. (A) Revised timeline for the CME pathway indicating the proposed temporal location of the cargo checkpoint. Proteins upstream and downstream of the checkpoint are listed under each phase. (B) Schematic cartoon illustrating the proposed mechanism leading to polarized endocytosis in budding yeast. In highly polarized cells, endocytic cargos (black arrows) are delivered primarily to the growing bud by exocytosis (gray arrows), locally accelerating the transition from the initiation and cargo-collection phase of CME (green spots) into the internalization phase (magenta spots). As exocytosis becomes depolarized later in the cell cycle, cargos are more plentiful across the plasma membrane, leading to global acceleration of maturation through the cargo checkpoint.
Paper by Michelle on a novel function of CDC42 published in JCB
Congratulations to Michelle on her paper “Cdc42 GTPase regulates ESCRTs in nuclear envelope sealing and ER remodeling ” now published in JCB.
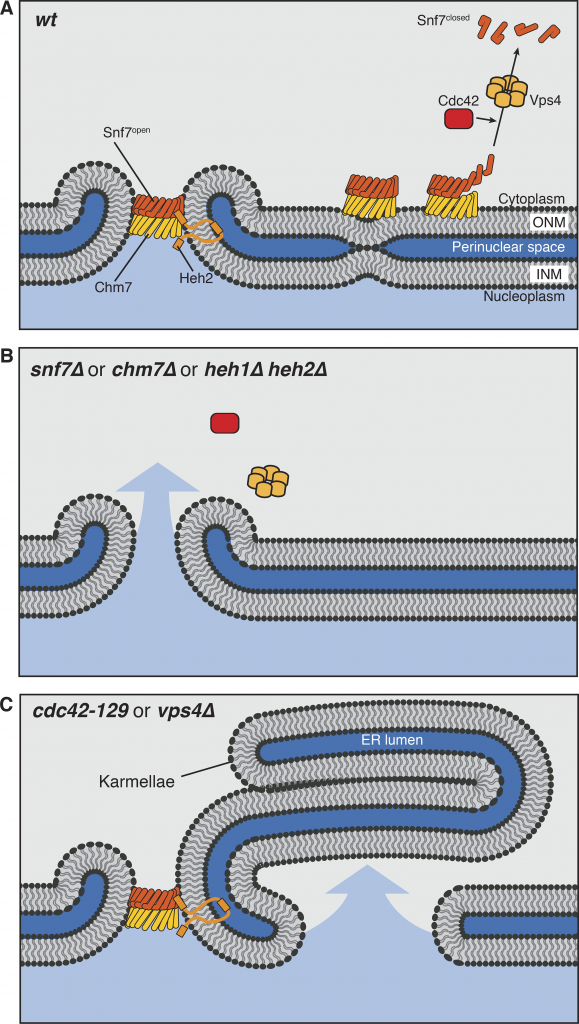
Cdc42, ESCRT-III, and Heh2 mutants share a leaky nucleus phenotype. (A) In normal cells, Heh2, Chm7, and Snf7 function at holes in the nuclear envelope to carry out annular fusion. ESCRT-III polymers are disassembled by Vps4, and we propose that Cdc42 is involved in the disassembly step by either directly contributing to Snf7 disassembly, activating Vps4 function, or cooperating with Vps4. (B) Mutants lacking components directly involved in annular fusion have holes in their nuclear envelopes left by nuclear fusion and ER fission events. (C) Cells lacking Vps4 and normal Cdc42 have unregulated ESCRT activity at the nuclear envelope. This causes the formation of nuclear karmallae and large holes in the nuclear envelope, leading to a defect in proper nucleo-cytoplasmic partitioning. ONM, outer nuclear membrane; INM, inner nuclear membrane.
Paper on combined SIM and AFM published in Nature Scientific Reports
Congratulations to former postdoc Daphne and collaborators in the Paiva and De Beule groups on their paper “Simultaneous co-localized super-resolution fluorescence microscopy and atomic force microscopy: combined SIM and AFM platform for the life sciences“, now published in the Nature Scientific Reports.
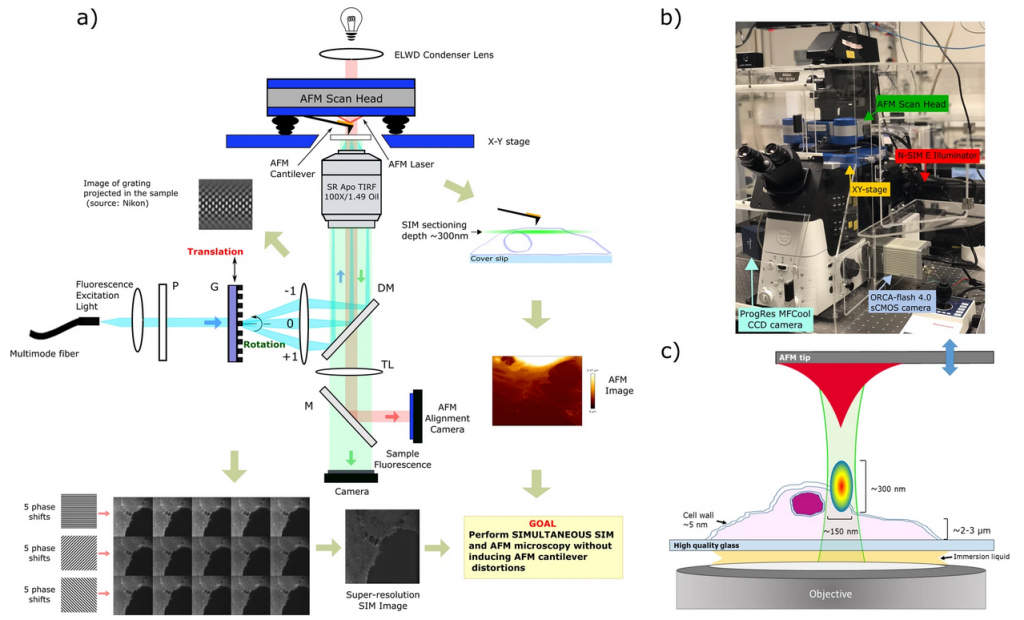
SR-SIM/AFM system.
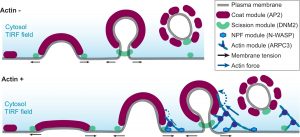
Paper by Matt on CME actin mechanics published in eLife
Congratulations to Matt, Daniel, Max, and our collaborators in the Rangamani Lab on their paper “Principles of self-organization and load adaptation by the actin cytoskeleton during clathrin-mediated endocytosis” now published in eLife.
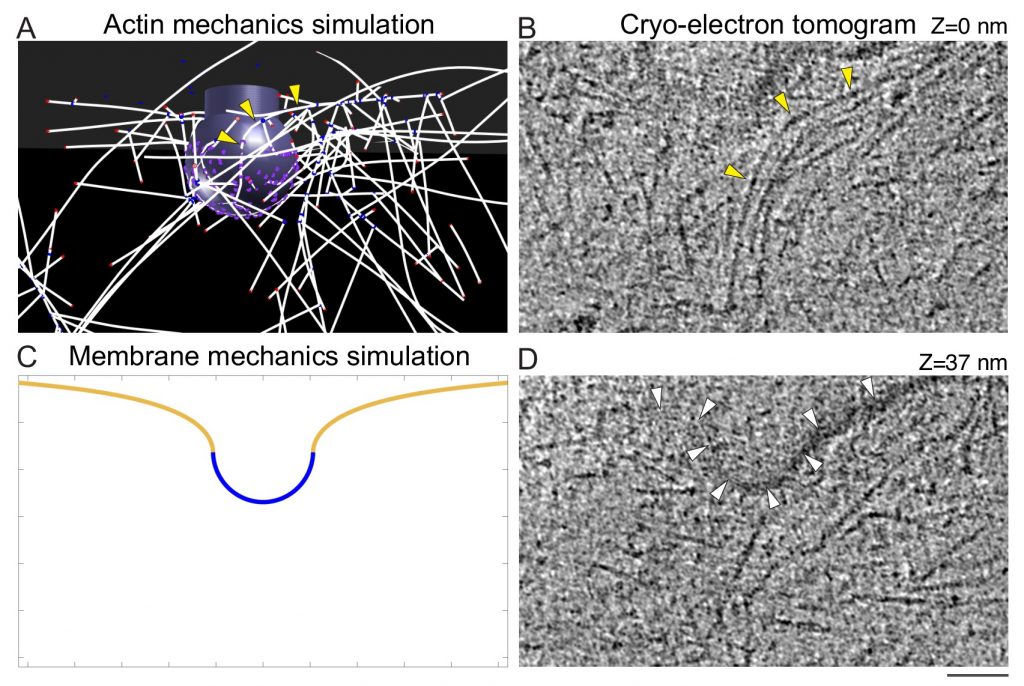
Multi-scale modeling predicts bent actin filaments during CME, as confirmed by cryo-electron tomography.