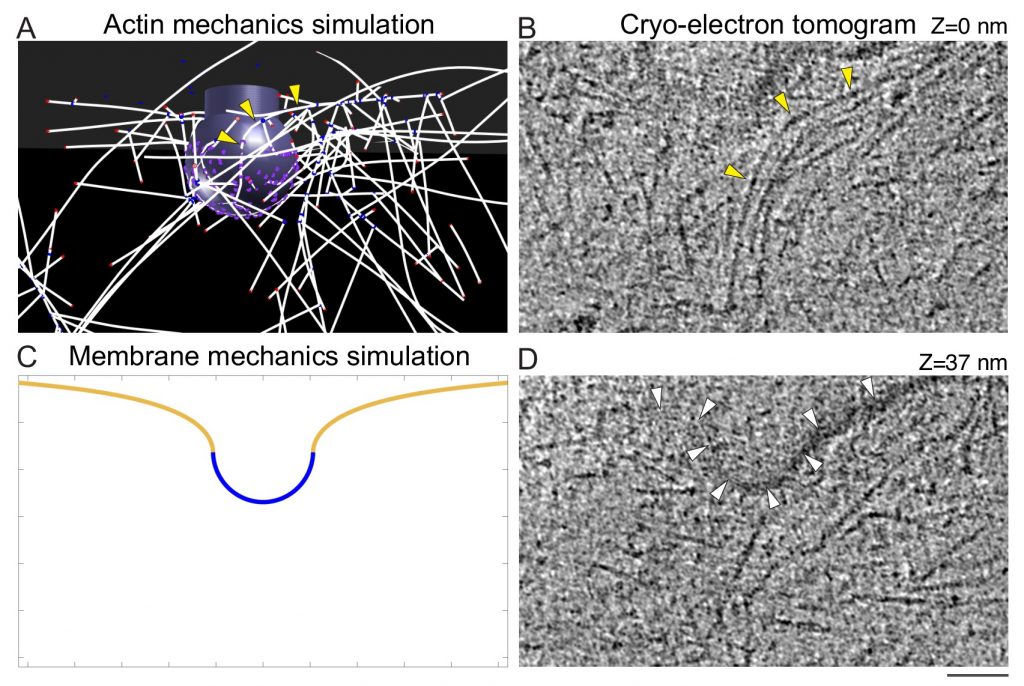
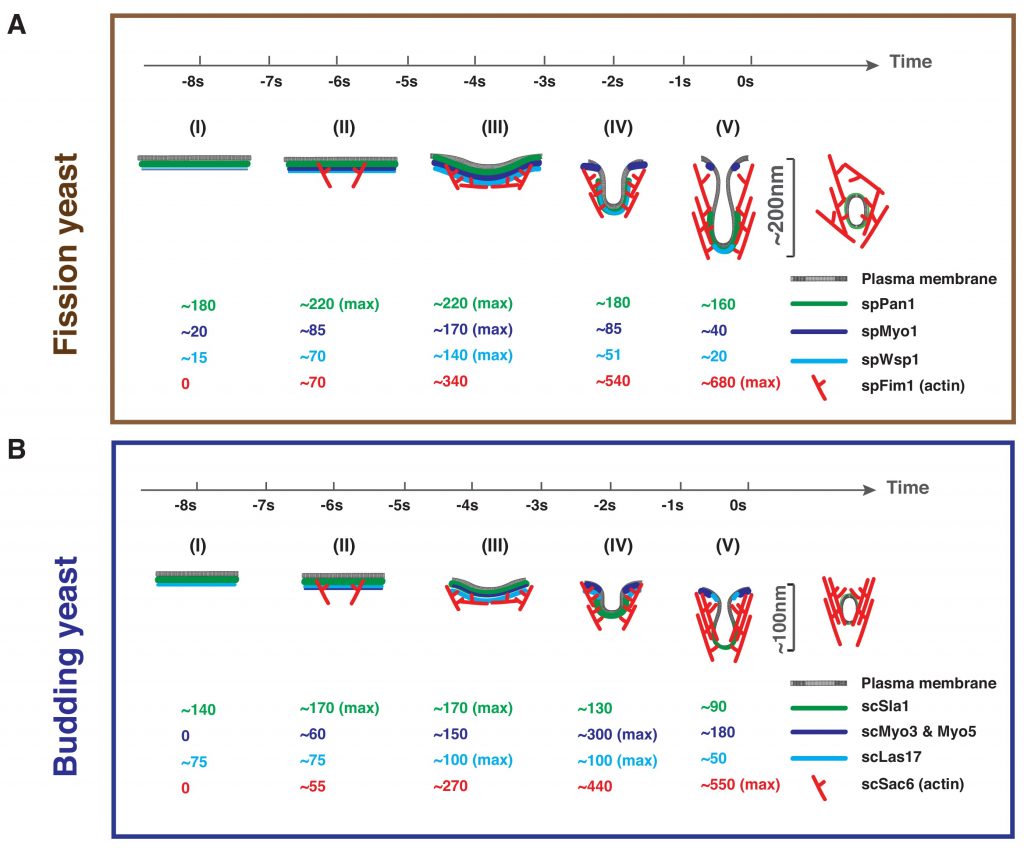
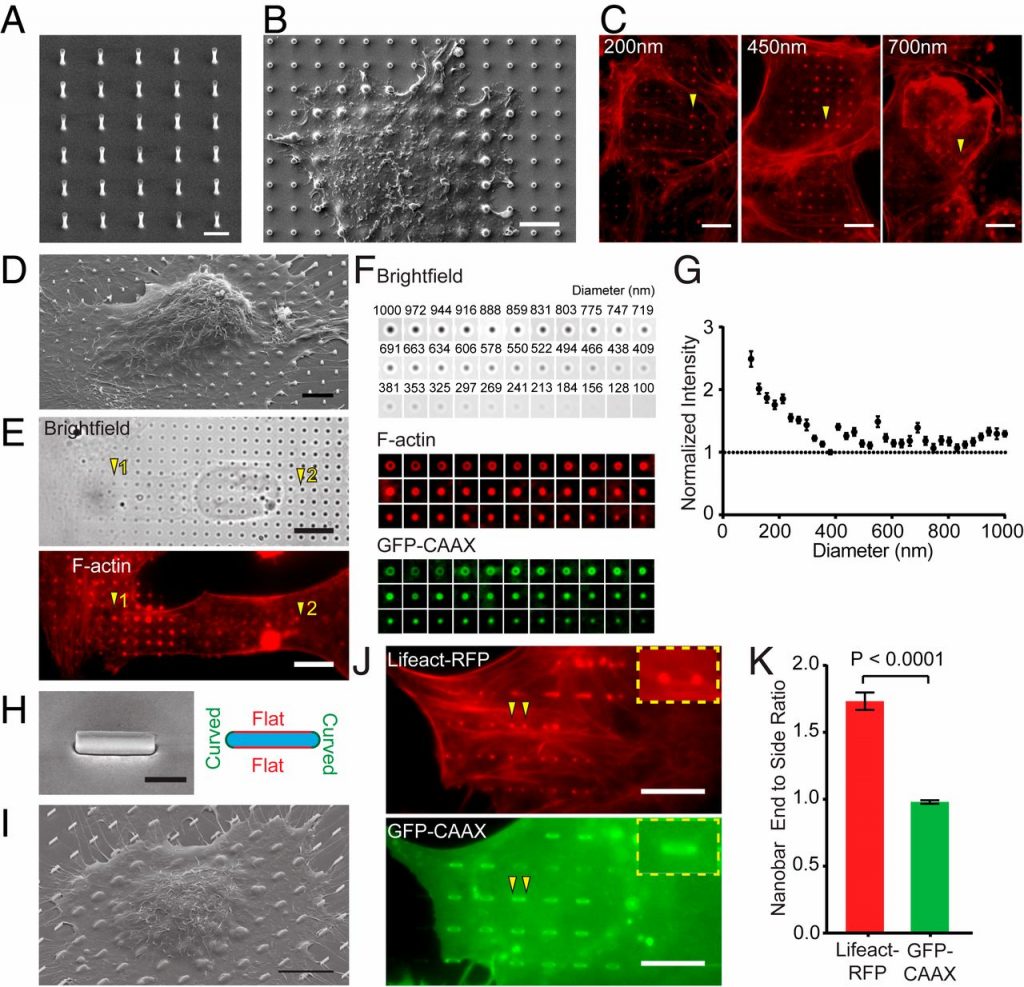
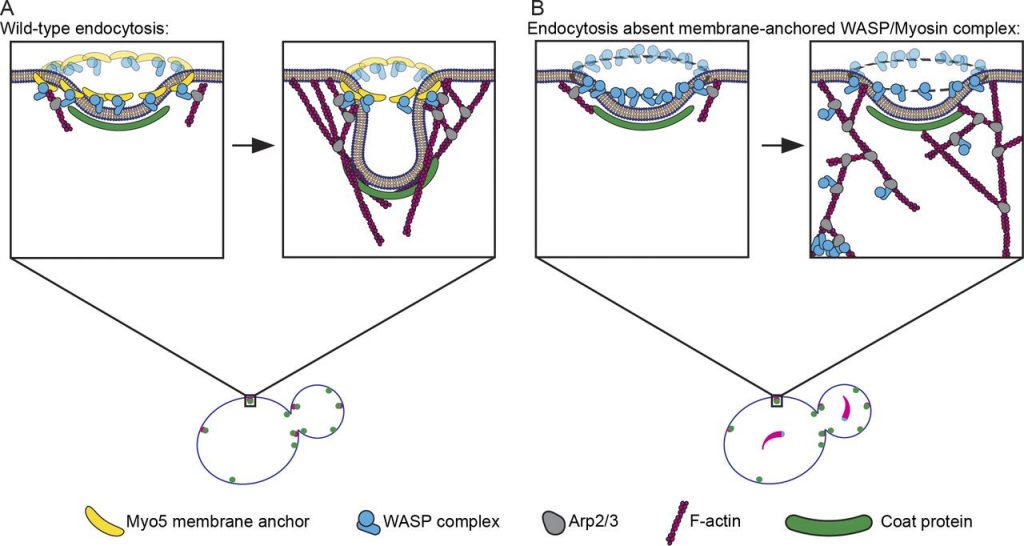
During clathrin-mediated endocytosis (CME), flat plasma membrane is remodeled to produce nanometer-scale vesicles. The mechanisms underlying this remodeling are not completely understood. The ability of clathrin to bind membranes of distinct geometries casts uncertainty on its specific role in curvature generation/stabilization. Here, we used nanopatterning to produce substrates for live-cell imaging, with U-shaped features that bend the ventral plasma membrane of a cell into shapes resembling energetically unfavorable CME intermediates. This induced membrane curvature recruits CME proteins, promoting endocytosis. Upon AP2, FCHo1/2, or clathrin knockdown, CME on flat substrates is severely diminished. However, induced membrane curvature recruits CME proteins in the absence of FCHo1/2 or clathrin and rescues CME dynamics/cargo uptake after clathrin (but not AP2 or FCHo1/2) knockdown. Induced membrane curvature enhances CME protein recruitment upon branched actin assembly inhibition under elevated membrane tension. These data establish that membrane curvature assists in CME nucleation and that the essential function of clathrin during CME is to facilitate curvature evolution, rather than scaffold protein recruitment.