Yansong Miao‘s new paper is out now at The Proceedings of the National Academy of Sciences. Congratulations to Yansong on his great work! The abstract is below. The PDF can be downloaded from PNAS here.
Cell-cycle regulation of formin-mediated actin cable assembly. Miao Y, Wong CC, Mennella V, Michelot A, Agard DA, Holt LJ, Yates JR 3rd, Drubin DG. Proc Natl Acad Sci U S A. 2013 Nov 19;110(47):E4446-E4455. Epub 2013 Oct 16. PMID: 24133141
Abstract
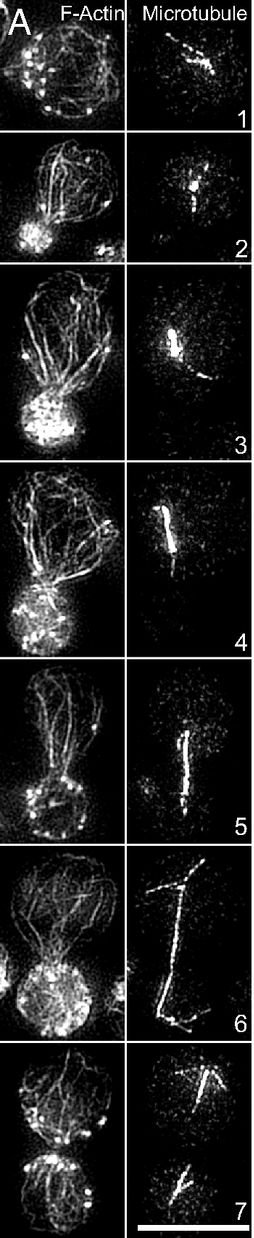
Assembly of appropriately oriented actin cables nucleated by formin proteins is necessary for many biological processes in diverse eukaryotes. However, compared with knowledge of how nucleation of dendritic actin filament arrays by the actin-related protein-2/3 complex is regulated, the in vivo regulatory mechanisms for actin cable formation are less clear. To gain insights into mechanisms for regulating actin cable assembly, we reconstituted the assembly process in vitro by introducing microspheres functionalized with the C terminus of the budding yeast formin Bni1 into extracts prepared from yeast cells at different cell-cycle stages. EM studies showed that unbranched actin filament bundles were reconstituted successfully in the yeast extracts. Only extracts enriched in the mitotic cyclin Clb2 were competent for actin cable assembly, and cyclin-dependent kinase 1 activity was indispensible. Cyclin-dependent kinase 1 activity also was found to regulate cable assembly in vivo. Here we present evidence that formin cell-cycle regulation is conserved in vertebrates. The use of the cable-reconstitution system to test roles for the key actin-binding proteins tropomyosin, capping protein, and cofilin provided important insights into assembly regulation. Furthermore, using mass spectrometry, we identified components of the actin cables formed in yeast extracts, providing the basis for comprehensive understanding of cable assembly and regulation.