Enjoy a video of the disco pumpkin made by Yidi Sun at the Drubin/Barnes, Koshland, Brar, Ünal, and Ingolia Labs (Barker Hall) pumpkin carving party to celebrate Halloween 2014:
Author Archives: NateKrefman
You can now follow David Drubin on Twitter @DavidGDrubin!
David Drubin, in his capacity as Editor-in-Chief of Molecular Biology of the Cell, has just joined Twitter. You can follow his tweets by clicking the link below:
Congratulations to Yansong Miao on his new paper!
Yansong Miao‘s new paper is out now at The Proceedings of the National Academy of Sciences. Congratulations to Yansong on his great work! The abstract is below. The PDF can be downloaded from PNAS here.
Cell-cycle regulation of formin-mediated actin cable assembly. Miao Y, Wong CC, Mennella V, Michelot A, Agard DA, Holt LJ, Yates JR 3rd, Drubin DG. Proc Natl Acad Sci U S A. 2013 Nov 19;110(47):E4446-E4455. Epub 2013 Oct 16. PMID: 24133141
Abstract
Assembly of appropriately oriented actin cables nucleated by formin proteins is necessary for many biological processes in diverse eukaryotes. However, compared with knowledge of how nucleation of dendritic actin filament arrays by the actin-related protein-2/3 complex is regulated, the in vivo regulatory mechanisms for actin cable formation are less clear. To gain insights into mechanisms for regulating actin cable assembly, we reconstituted the assembly process in vitro by introducing microspheres functionalized with the C terminus of the budding yeast formin Bni1 into extracts prepared from yeast cells at different cell-cycle stages. EM studies showed that unbranched actin filament bundles were reconstituted successfully in the yeast extracts. Only extracts enriched in the mitotic cyclin Clb2 were competent for actin cable assembly, and cyclin-dependent kinase 1 activity was indispensible. Cyclin-dependent kinase 1 activity also was found to regulate cable assembly in vivo. Here we present evidence that formin cell-cycle regulation is conserved in vertebrates. The use of the cable-reconstitution system to test roles for the key actin-binding proteins tropomyosin, capping protein, and cofilin provided important insights into assembly regulation. Furthermore, using mass spectrometry, we identified components of the actin cables formed in yeast extracts, providing the basis for comprehensive understanding of cable assembly and regulation.

Happy Halloween from the Drubin/Barnes Lab!!
Microtubules and Mitosis Journal Club on Monday, August 26
For Microtubules and Mitosis Journal Club on August 26, members of the Drubin/Barnes Lab, Eva Nogales Lab, and Ahmet Yildiz Lab will discuss the following paper, selected by Itziar Ibarlucea:
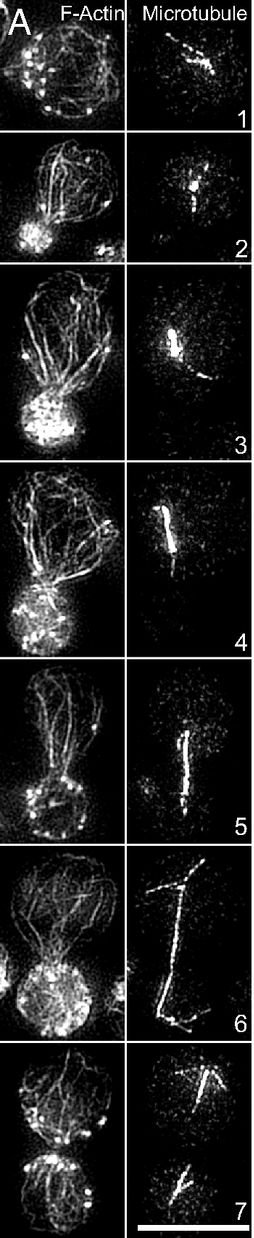
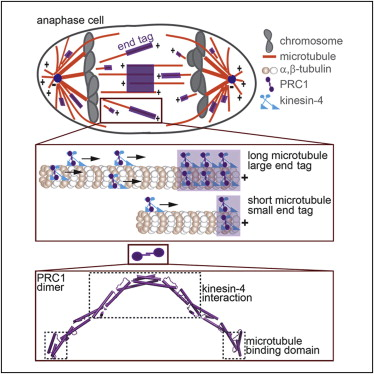
Marking and Measuring Single Microtubules by PRC1 and Kinesin-4. Subramanian R, Ti SC, Tan L, Darst SA, Kapoor TM. Cell. 2013 Jul 18;154(2):377-90. PMID: 23870126.
Journal Club on Monday, August 6
For Journal Club on August 6, Nate Krefman presented the following paper:
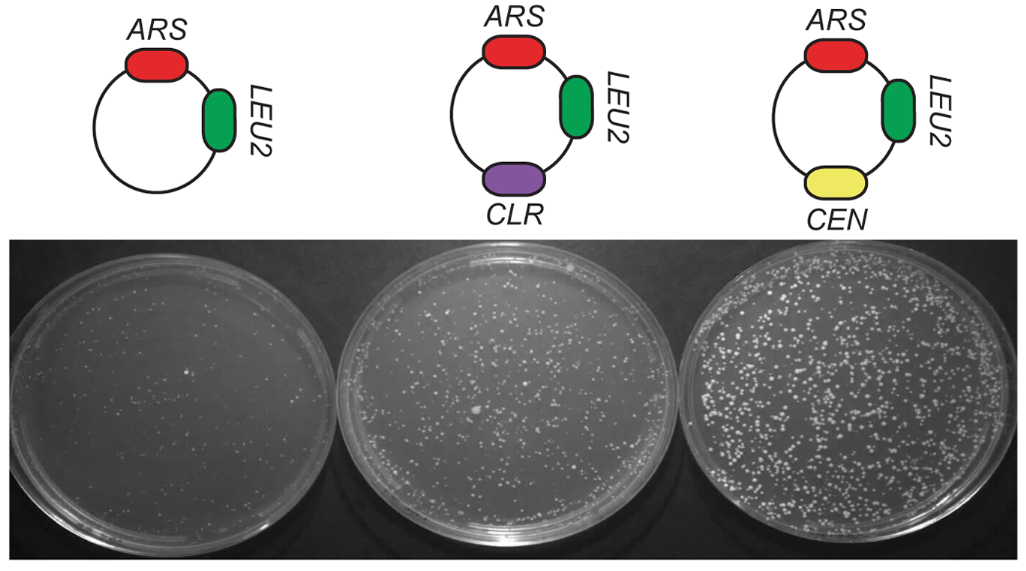
Centromere-like regions in the budding yeast genome. Lefrançois P, Auerbach RK, Yellman CM, Roeder GS, Snyder M. PLoS Genet. 2013;9(1):e1003209. PMID: 23349633.
Journal Club on Monday, August 6
For Journal Club on August 6, Eric Lewellyn presented the following paper:
Actin filament severing by cofilin dismantles actin patches and produces mother filaments for new patches. Chen Q, Pollard TD. Curr Biol. 2013 Jul 8;23(13):1154-62. PMID: 23727096.
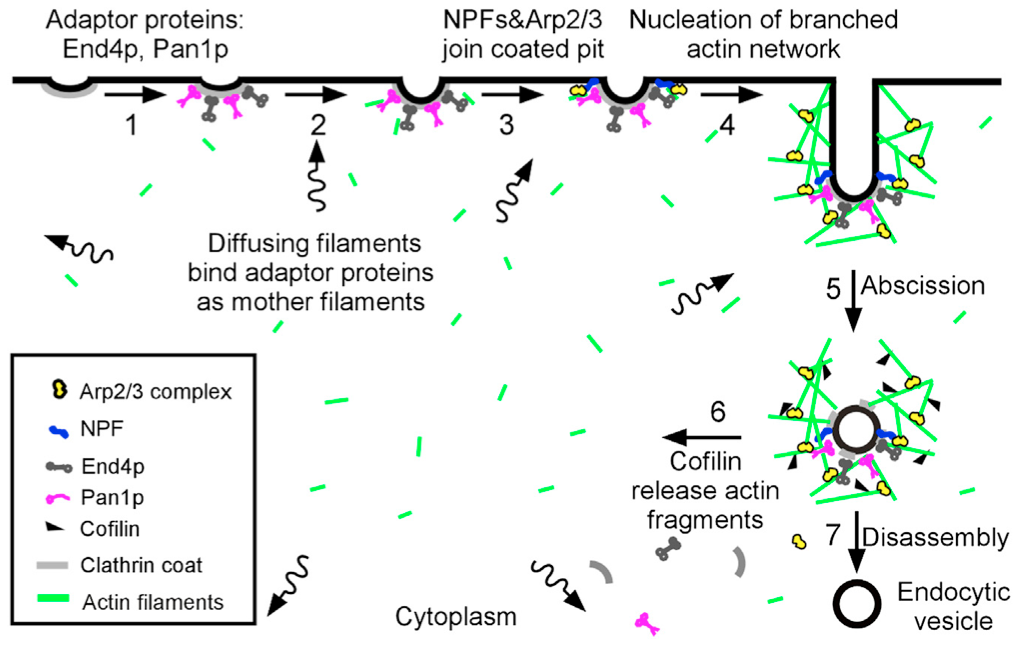
“Sever, diffuse and trigger” model for actin filament turnover in actin patches. The 7 steps (numbers next to the arrows) are (1) clathrin coated pits bind adaptor proteins End4p and Pan1p, (2) short, diffusing actin filaments bind to End4p and Pan1p associated with coated pits, and (3) Arp2/3 complex interacts with these mother filaments and nucleation promoting factors to (4) initiate branching nucleation of actin filaments that promote elongation of the endocytic tubule. (5) After abscission of the vesicle, (6) cofilin severs actin filaments to generate a pool of short, diffusing actin filaments, some of which return to the cycle at step 2.
Journal Club on Monday, July 22
For Journal Club on July 22, Connie Peng presented the following paper:
Mps1 and Ipl1/Aurora B act sequentially to correctly orient chromosomes on the meiotic spindle of budding yeast. Meyer RE, Kim S, Obeso D, Straight PD, Winey M, Dawson DS. Science. 2013 Mar 1;339(6123):1071-4. PMID: 23371552
Microtubules and Mitosis Journal Club on Monday, June 24
For Microtubules and Mitosis Journal Club on April 25, members of the Drubin/Barnes Lab, Eva Nogales Lab, and Ahmet Yildiz Lab will discuss the following paper, selected by Stu Howes:
Synergy between XMAP215 and EB1 increases microtubule growth rates to physiological levels. Zanic M, Widlund PO, Hyman AA, Howard J. Nat Cell Biol. 2013 Jun;15(6):688-93. PMID: 23666085.
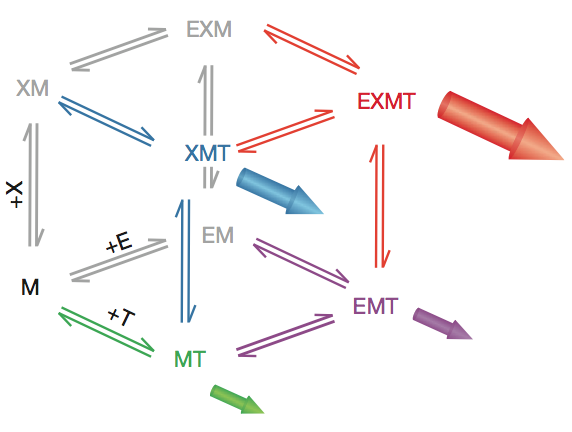
Reaction scheme for an enzymatic reaction with two non-essential activators. The microtubule end is viewed as an enzyme that catalyses the reaction: tubulin in solution (substrate)→tubulin in the polymer (product) by providing a much more efficient pathway than direct incorporation into the lattice. XMAP215 and EB1 are viewed as two non-essential enzyme activators that accelerate the net rate of addition at the end. Filled arrows represent the flux of polymerization through a particular vertex. M, microtubule end; T, free tubulin; X, XMAP215; E, EB1.
Journal Club on Monday, June 17
For our Journal Club on June 17, Jessica Marks will present the following paper:
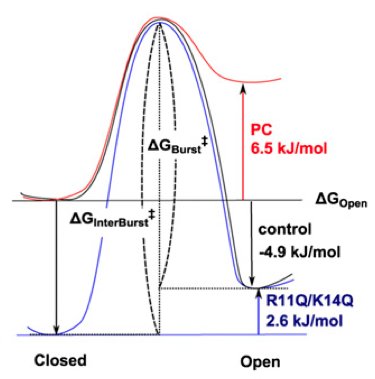
Amphipathic antenna of an inward rectifier K+ channel responds to changes in the inner membrane leaflet. Iwamoto M, Oiki S. Proc Natl Acad Sci U S A. 2013 Jan 8;110(2):749-54. PMID: 23267068.